Contents
Oligonucleotides as drugs
Introduction
Traditional small-molecule antiviral agents inhibit enzymes that are important to viral replication or reverse transcription. The synthesis and design of nucleoside-based antiviral agents is well-established and many clinically important drugs have been developed (e.g. acyclovir and AZT). However, it has proved difficult to develop compounds with the ability to eliminate viruses entirely or to prevent their integration into the host genome. For this reason a number of oligonucleotide-based approaches are now being developed.
Antisense oligonucleotides
Viruses and bacteria have unique proteins that are essential for their survival. The mRNA sequences that encode these proteins are absent in the human genome, so any method to eliminate foreign mRNA molecules without interfering with the mRNA of the host is potentially of great therapeutic value. Moreover, advances in DNA sequencing technology have made the information required to target pathogens readily obtainable. Antisense oligonucleotides are designed to do precisely this job. They hybridize to specific mRNAs and inhibit expression of particular proteins.
If the protein in question is important for the replication of a virus, or for the uncontrolled growth of cancer cells, a therapeutic effect may be observed. In theory, antisense oligonucleotides can also be used to block the synthesis of proteins in normal cells so that the biological effect of specific proteins can be determined. However, in this application the recently developed RNAi method is more popular. The RNA:DNA hybrid duplex that is formed between the antisense oligonucleotide and the target mRNA interferes directly with protein synthesis, resulting in reduced expression of the coded protein. This inhibition can be caused by the inability of the ribosome to assemble around the mRNA when it is bound to the antisense oligonucleotide and/or the inability of the hybrid to pass through the ribosome and direct protein expression. The antisense effect is enhanced by the endonuclease activity of ribonuclease H (RNase H) on mRNA molecules that are hybridized to antisense oligonucleotides. RNase H destroys the mRNA strand, and both the RNAse H enzyme and the antisense oligonucleotide remain intact at the end of the cycle: the antisense effect is therefore catalytic (Figure 1). A single antisense oligonucleotide can partake in the destruction of many mRNA molecules by this mechanism. The enzyme RNase H occurs in normal cells where it plays an important role in removing short pieces of RNA from the lagging strand of DNA replication.
There are 3 × 109 base pairs in the human genome. Therefore, statistically any DNA sequence of 17 bases or more will be expected to occur only once or not at all (417 is greater than 1010). If a region of the mRNA of a viral protein is targeted with an antisense oligonucleotide the oligonucleotide should be selective for the viral mRNA.
DNase-resistant antisense oligonucleotides
Unfortunately, simply adding chemically unmodified antisense oligonucleotides to cells in culture or to organisms in vivo does not normally result in a significant antisense effect. This is because normal unmodified oligonucleotides are unstable in biological media and are degraded by endogenous DNase enzymes. It is therefore necessary to chemically modify antisense oligonucleotides to make them stable in cells. There are different types of DNase enzymes: those with 5'-exonuclease activity digest the oligonucleotide from the 5'-end; 3'-exonucleases digest from the 3'-end; and endonucleases digest from within the DNA chain. Whatever the nature of the DNase enzyme, any modification of the phosphodiester backbone is likely to inhibit its action and several phosphodiester backbone analogues have been developed with this goal in mind (Figure 2).
Some modified phosphodiester backbones, such as methyl phosphonates, are less desirable because they prevent the mRNA of the DNA:mRNA hybrid from being a substrate for RNase H. Methyl phosphonates also have a detrimental effect on the aqueous solubility of the antisense oligonucleotides because the backbone is uncharged. The phosphorus atoms in the backbone of phosphorothioate and methyl phosphonates are chiral and the methods of synthesizing these analogues are not stereospecific. Therefore, the resulting oligonucleotides are mixtures of diastereomers (Figure 3). This is not an ideal situation from a pharmaceutical standpoint, but it has not prevented regulatory bodies from allowing antisense oligonuclotides to be used in the clinic.
The duplexes formed between backbone-modified antisense oligonucleotides and target mRNAs are generally thermodynamically less stable than normal DNA:RNA hybrids. In the case of phosphorothioates this is due to the steric bulk of the sulfur atoms which interferes with duplex formation.
Phosphorothioate oligonucleotides are synthesized by the solid-phase phosphoramidite method using tetraethylthiuram disulfide (disulfiram) to introduce the sulfur atom instead of the iodine oxidation step (Figure 4). Because SODNS are required in large quantities for clinical studies, solid-phase methods have been developed to synthesize them in multi-kilogram quantities. Methyl phosphonate oligonucleotides are also synthesized by the phosphoramidite method using methyl phosphonamidites (Figure 5). The methyl phosphonate group is not stable to aqueous base, so the protecting groups on the heterocyclic bases have to be cleavable under mild conditions.
In the antisense field more work has been carried out on phosphorothioate oligonucleotides (SODNS) than on any other modification. This is because SODNS are easy to synthesize, have good resistance to undesirable DNase degradation and do not inhibit the beneficial RNase H digestion of mRNA in SODN:mRNA hybrids.
Modifications to the sugar moiety of antisense oligonucleotides, which also prevent degradation by DNase enzymes, are summarized in Figure 6. It is possible to combine more than one modification in a single oligonucleotide: one common strategy is to alternate stretches of modified backbone with lengths of normal (phosphodiester) DNA backbone. For example oligonucleotides with phosphorothioate or methyl phosphonate groups at the 5'- and 3'-ends tend to have reasonably good stability in vivo, presumably because DNA exonucleases are more prevalent in cells than endonucleases.
Cellular uptake
Oligonucleotides have a much greater molecular mass than conventional small-molecule pharmaceutical drugs, and this is one reason why their cellular uptake by passive diffusion is inefficient. Fortunately, some cells take in oligonucleotides by a process known as receptor-mediated endocytosis, which involves the specific uptake of the oligonucleotide by natural cell-surface receptors. SODNS are taken up reasonably efficiently by this process. During the process of cell uptake oligonucleotides are sometimes trapped in endosomes and separated from target mRNA by an impermeable membrane. Chemically unmodified oligonucleotides are rapidly digested in endosomes but some chemically-modified antisense oligonucleotides survive long enough for the endosomes to break down and release their contents into the cytoplasm.
Non-antisense effects of therapeutic oligonucleotides
Some oligonucleotides do not exert their therapeutic effect by the predicted antisense mechanism but by stimulating the natural immune response of the host cell. Oligonucleotides with sequences containing one or more CpG (CG) units can bind to the protein TLR9, and this triggers the induction of cell signalling pathways that lead to the stimulation of the cell's innate immune response.
The triplex approach to gene inhibition
The antisense approach is elegant, but suffers from one major potential disadvantage: there are often very large numbers of mRNA molecules in a cell, and it is is therefore difficult to achieve complete inhibition of a specific mRNA. Moreover, feedback mechanisms exist that can lead to increased mRNA production in response to destruction of mRNA in antisense therapy. In principle a much more attractive approach to blocking the synthesis of specific proteins is direct inhibition of genomic DNA. There are only two copies of genetic information in the nucleus of the human cell, and, even in the case of genes that occur in tandem, only a handful of copies normally exist. Switching off genes directly by an external agent is an extremely attractive proposition and in principle it could be achieved by sterically blocking the double helix so that the proteins that interact with DNA can no longer bind. This would directly prevent replication and transcription, the mechanisms by which DNA and the RNA message are copied. It would also offer the possibility of interfering with regions of DNA involved in the regulation of gene expression. The challenge is to develop a chemical agent that can bind tightly to a specific region of duplex DNA in the presence of the entire human genome. This is a major task but it is not insurmountable. If such an agent could be developed it would have profound implications in molecular biology, diagnostics and in medicine.
Nature offers us clues to solving the problem. It has been known for some time that a third strand of natural DNA can fit into the major groove of the DNA duplex and interact with the base pairs to form a triple helix (Figure 7).
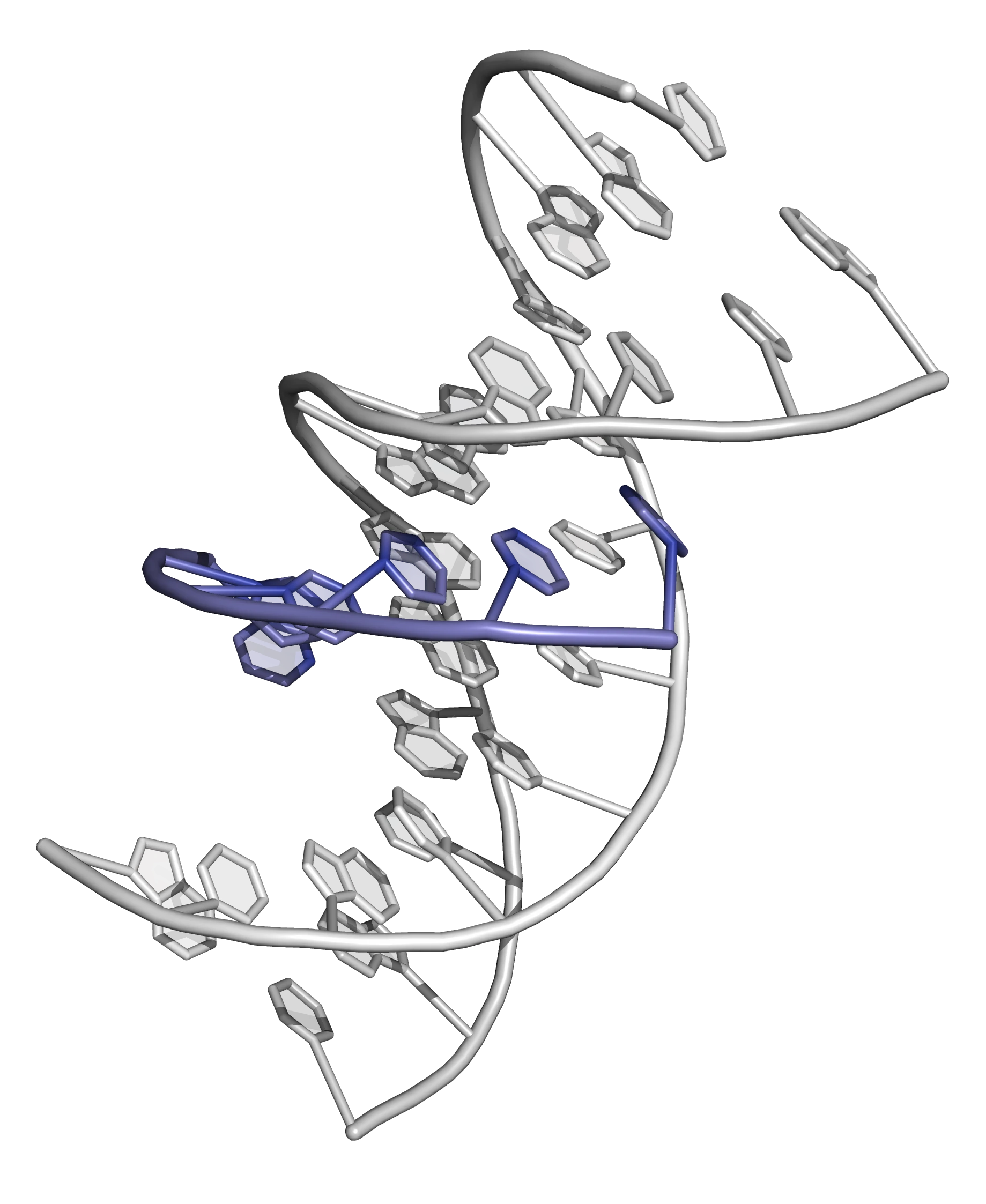
As each base pair in DNA (A·T, T·A, G·C and C·G) has a unique profile in the major groove it should be possible to specifically recognize (and "read") any DNA sequence by probing this groove (Figure 8).
Triple helices were first observed by Felsenfeld, Davies and Rich in 1957. However, using natural DNA or synthetic oligonucleotides containing natural bases it was only possible to create triplexes if one of the strands of the duplex was purine-rich.
Triple helices are able to form because the purine bases of DNA have hydrogen bonding donors and acceptors that are capable of forming two additional hydrogen bonds with bases in the major groove. These bonds are called Hoogsteen bonds. Duplexes capable of forming triple helices contain purine bases in one strand and pyrimidines in the complementary strand, so a pre-requisite for triplex formation is a homopyrimidine/homopurine region of DNA. Triplex forming oligonucleotides (TFOs) can be oriented antiparallel or parallel to the purine strand of the duplex Figure 9).
Antiparallel triplexes are intrinsically unstable at all pH values. The TFO is composed mostly of G and A bases. Antiparallel triplets are shown in Figure 10.
In the parallel orientation a third strand thymine recognizes A·T and protonated cytosine recognizes G·C (Figure 11).
Parallel triplexes are thermodynamically stable at room temperature below pH 6 in aqueous buffer, but are less stable at physiological pH. There are two reasons for the instability of triplexes at pH 7 and above:
- the C+·GC triplet is stabilized by protonation and this only occurs at low pH, as the pKa of N(3) of cytosine is 4.5;
- the juxtaposition of three polyanionic DNA strands is destabilizing at all pH values.
In addition to the low affinity of the TFO for the DNA duplex, the requirement for polypurine:polypyrimidine regions is a major restriction in terms of target selection, as there are few naturally occurring DNA sequences of this kind.
An important potential application of triple helixes is the use of single-stranded oligonucleotides to target genomic DNA, interfere with replication and transcription and thereby destroy cancer cells or viruses. However, before such a therapeutic regime can be contemplated there is a need to develop triplexes that are both stable and sequence-specific under physiological conditions.
Chemically modified triplex-forming oligonucleotides
Chemists are currently addressing the triplex stability problem in an effort to develop triplex-forming oligonucleotides (TFOs) for commercial use in genomics and diagnostics as well as biomedical applications. Triple helix stability can be improved by incorporating modified nucleosides into the TFO and some modified bases that have been developed to improve triplex stability and sequence recognition are shown in Figure 12. Some important examples are
- the anilinothiazole S that specifically binds to T·A base pairs by H-bonding to the T and A bases;
- the methyl pyrimidone base PO that has been used to stabilize triplexes which contain C·G base pairs by the formation of an N-H…N hydrogen bond and a weak C-H...O hydrogen bond;
- the pyrimidine analogue MP (3-methyl-2-aminopyridine) that specifically recognizes G·C. It has a pKa > 6, and unlike cytosine it is partially protonated at physiological pH.
- the 5-aminopropargyl-2'-aminoethoxythymidine nucleoside Ba which has two primary amines that are protonated at pH 7, and interact with the anionic phosphate groups in the neighbouring duplex. The triple bond stabilizes the correct helical conformation in the third strand through stacking interactions with the surrounding bases (base-stacking). It is important that the TFO is able to adopt a stable conformation that is compatible with the shape of the major groove of the duplex. These chemical modifications go some way towards solving the "triplex stability problem" and it is likely that further advances will be made in the near future.
Triplex-forming oligonucleotides labelled with intercalators
Labelling TFOs with an intercalative fluorophore such as thiazole orange can increase the stability of the triplex at neutral pH. Intercalators improve the stability of both duplexes and triplexes by inserting themselves in between adjacent base pairs of DNA (forming energetically favourable stacking interactions).
Tethering an intercalator to a parallel TFO can be achieved using post-synthetic labelling of an oligonucleotide containing an amino-modified nucleobase such as 5-propargylamino-dT (Figure 13). As a result, very stable triplexes are obtained at (and above) neutral pH, and the stabilizing effects of the intercalator are cumulative: three additions of thiazole orange can increase the melting temperature of the triplex by up to 50 °C, with a corresponding 1000-fold improvement in Kd. This greatly increased stability expands the range of target duplexes at neutral pH to include polypurine sequences that contain a pyrimidine inversion such as CG. This approach could unlock many practical applications of triplexes and might also be effective in the stabilization of highly unstable antiparallel triplexes.
In addition to its stabilizing behaviour, thiazole orange exhibits a large increase in fluorescence on binding to double-stranded DNA. This makes thiazole orange-labelled TFOs potentially useful as probes for detection of genomic sequences.
RNA Interference (RNAi)
The under- or overexpression of a protein, or expression of a protein from a mutated, viral or bacterial gene, can lead to the occurrence of disease in humans. The messenger RNA of these proteins represents an excellent therapeutic target, and interfering with mRNA expression can prevent the production of undesirable proteins without destroying the genetic material.
RNA interference (RNAi), a natural process that occurs when double-stranded RNA is introduced into cells, causes sequence-specific gene silencing. RNAi was first observed in transgenic plants when, in an effort to increase flower pigmentation in petunias, a transgene (chalcone synthase) was introduced. The resulting plants had variegated petals, indicating that gene expression had been affected. Researchers soon discovered that injecting double stranded RNA (dsRNA) into cells led to efficient sequence-specific gene silencing. It was deduced that the mechanism involved cleavage of these long dsRNAs into short dsRNA species (small interfering RNAs; siRNAs), caused by a dsRNA-specific endoribonuclease called Dicer. It was also observed that introduction of chemically synthesized 21-23 nucleotide siRNAs also caused degradation of homologous RNA.
The accepted mechanism by which RNAi silences genes is outlined in Figure 14. Long, double-stranded RNA (typically >200 nucleotides) is cleaved by the RNA III protein Dicer into short RNA duplexes (siRNA) in an ATP-dependent reaction. Typically the siRNAs (21-23 nucleotides in length) are phosphorylated at the 5'-end and have two unpaired nucleotides at their unphosphorylated 3'-ends. The siRNAs are assembled with cellular proteins to form the RNA-induced silencing complex (RISC) that contain endoribonucleases. The siRNA is unwound and the single-stranded antisense strand directs the RISC, by the specificity of base-pairing, to a target messenger RNA (mRNA). This results in endonucleolytic cleavage of the mRNA at a specific site in the centre of the duplex, and thereby silences the gene in a sequence-specific manner.
Introduction of dsRNA (> 30 nucleotides) into mammalian cells can induce a potent antiviral response causing non-specific inhibition of protein synthesis and degradation of RNA. To avoid this, siRNAs are either introduced or expressed directly.
RNAi is an important technique as it can be used to identify the functions of specific genes. It could also be used to develop a new class of drugs that offer high specificity and a rapid response. The drugs may be engineered to selectively silence target genes, permanently switching them off, and reduce or avoid side-effects associated with conventional medicines.
DNA Quadruplexes
The ends of chromosomes (telomeres) consist of highly repeated G-rich sequences. In humans the sequence is (GGGTTA)n where n is several hundred. These sequences cannot be fully copied as it is not possible for replication to extend right to the end of a DNA duplex, so the telomeres get progressively shorter through multiple cycles of cell division. Eventually the number of (GGGTTA)n repeats falls below a critical level and the cell dies. This is probably a process that has evolved to define the useful lifetime of a cell. However, in stem cells and in most cancer cells, (GGGTTA)n repeats are added to the ends of chromosomes by a specific enzyme called telomerase, which is not present in most normal differentiated cells and the cells become immortal. This means that telomerase may be a good target for novel antitumour agents. Telomerase incorporates a single strand of RNA containing the sequence rUUACCC which acts as a template for the synthesis of the dGGGTTA repeats.
It is known that G-rich sequences such as telomeres can fold into quadruplexes (Figure 15; Figure 16). These structures form slowly, but are extremely stable, with high melting temperatures.
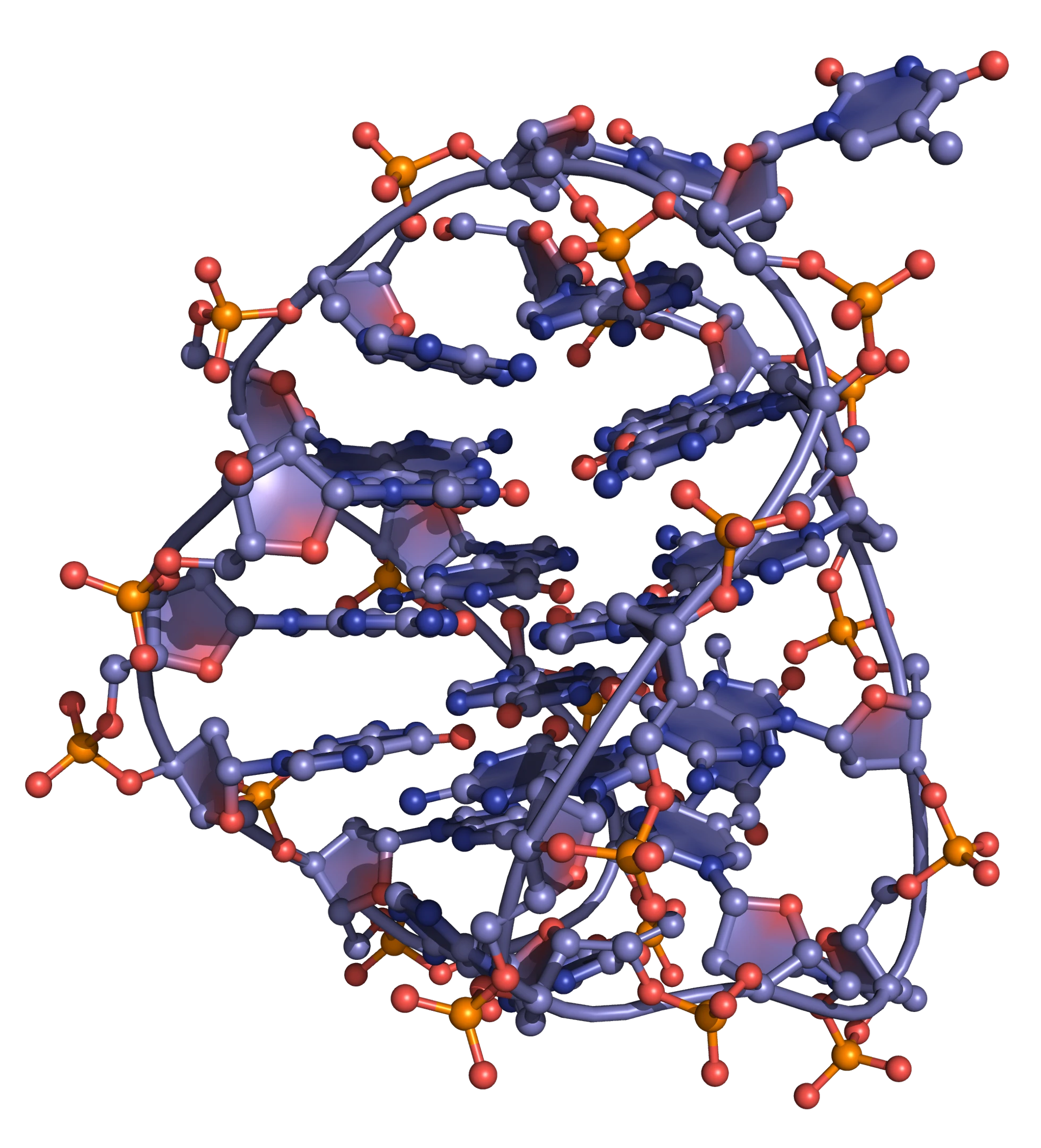
Quadruplexes consist of stacks of G-quartets, formed by the association of four G-rich DNA strands and stabilized by metal ions (commonly potassium, but occasionally sodium; Figure 17). These "four strands" can actually be part of the same single strand of DNA: in this case the quadruplex is intramolecular (or unimolecular).
It has not been proved that telomeres exist as quadruplexes in vivo but it is possible that these structures have some biological role. Quadruplexes are interesting from a structural point of view as different arrangements of the strands can give rise to alternative quadruplex structures (Figure 15).
There has been substantial recent work on developing drugs that bind to DNA quadruplexes. Molecules that can either facilitate quadruplex formation or stabilize quadruplexes have the potential to interfere with replication, inhibit telomerase activity and arrest cancer cell growth.